NTU_Taiwan
Our Project

Motivation

According to the World Health Organization, lung cancer has the highest cancer mortality rate worldwide. The main cause is gene mutation or abnormal differentiation of lung cells, which causes the cells to lose their normal growth regulation ability, and then proliferate and divide without limit, forming malignant tumors (WHO, 2023). Lung cancer refers to any malignant tumor that develops in the lungs, trachea, or bronchi, depending on the site of origin. Risk factors for lung cancer vary, including smoking, long-term exposure to high concentrations of carcinogens, family history, and chronic lung disease.
Currently, commonly used clinical treatments include surgical resection, radiotherapy,
chemotherapy, targeted drug therapy, and immunotherapy. Among them, traditional chemotherapy
prevents tumor proliferation and metastasis by inhibiting rapid cell division,
but due to its lack of selectivity, it often damages normal cells at the same time,
leading to serious side effects. Targeted drug therapy, which has emerged in recent years,
can target specific mutated genes (such as EGFR、ALK、ROS1) to inhibit, which is specific and
reduces side effects (Yuan M., Huang L-L., Chen J-H., Wu J., & Xu Q., 2019). However, there are
still many problems with this type of treatment limit:
- Tumor cells are prone to secondary mutations, which lead to changes in drug binding sites and drug resistance.
- Targeted drugs are only available for patients with specific mutations, and more than 70% of lung cancer patients cannot benefit from existing drugs.
- Targeted therapy lacks long-term immune memory, and the disease is prone to recurrence once the drug is stopped or the tumor escapes mutation.
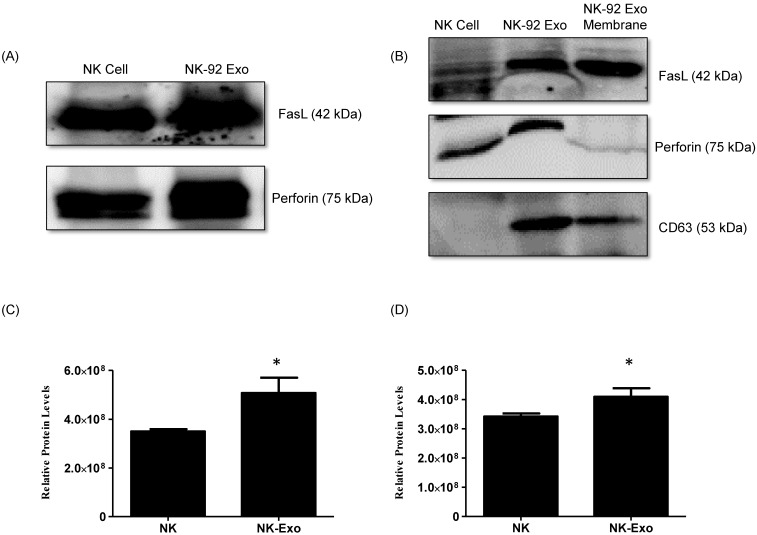
However, exosomes have limited directional properties, making it difficult to accurately deliver cargo to specific target cells, affecting the treatment effect. Therefore, we use DNA origami. The structure, used as a high-precision nanocarrier combined with NK92-derived exosomes, can improve the targeting of lung cancer cells and then use the ability of exosomes to kill cancer cells. It is hoped that DNA origami can be used to establish a nano-delivery system with targeted and immune-modulating potential.

Design

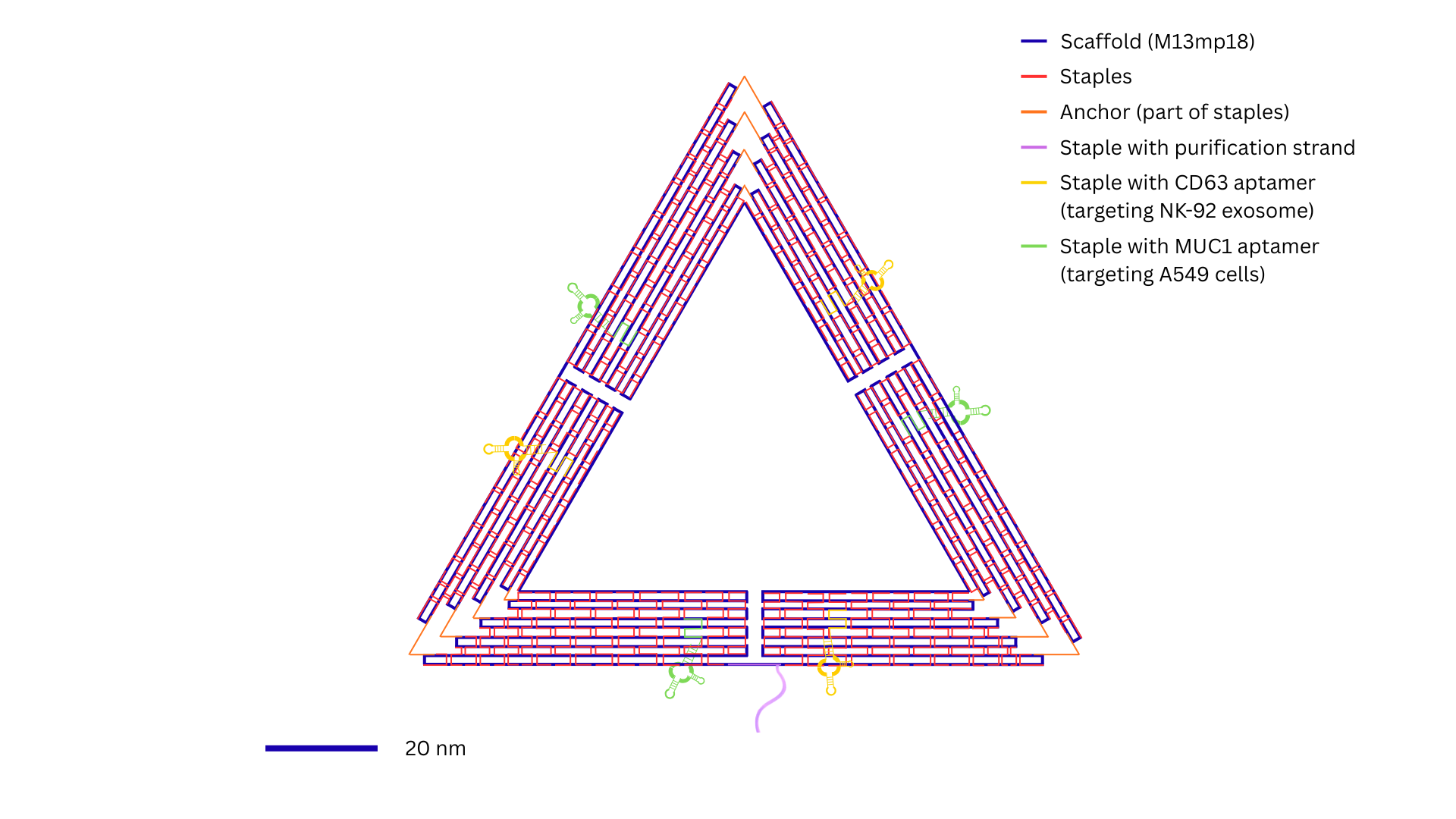
In addition to the staple strands, three kind of
specific DNA sequences were introduced: CD63 aptamer (targeting NK-92-drived exosomes)
(Zhu, L., Kalimuthu, S., Gangadaran, P., Oh, J. M., Lee, H. W., Baek, S. H., Jeong, S. Y., Lee,
S. W., Lee, J., & Ahn, B. C., 2017) , a MUC1 aptamer (targeting A549 cells) (Shahrad, S., Rajabi,
M., Javadi, H., Karimi Zarchi, A. A., & Darvishi, M. H., 2022), and a DNA oligonucleotide designed
for magnetic bead-based purification (Jingjing Ye, Josephine Teske, Ulrich Kemper, & Ralf Seidel., 2021).
The DNA origami folding reaction was performed in 1× TAE buffer supplemented with 10 mM MgCl2,
using a scaffold-to –staple molar ratio of 1:10. Thermal annealing was carried out by heating the mixture to
80 °C, and subsequently cooling it to room temperature over
a time course of 90 min to promote proper folding. Subsequently, the functional DNA fragments
(added at the same molar concentration and volume as the staple strands) were incubated with the
pre-folded structure at 37°C for 2 hours to facilitate hybridization at designated overhand sites
(Rothemund, P., 2006; Kielar, C., Yang, X., Xu, X., Zhu, S., Gorin, N., Grundmeier, G., Möser, C.,
Smith, D. M., & Keller, A., 2019).
The resulting structure consisted of a triangular DNA origami plane bearing seven single-stranded
overhangs. Transmission electron microscopy (TEM) imaging confirmed the formation of uniform,
equilateral triangular nanostructures, consistent with the origami design created in caDNAno.
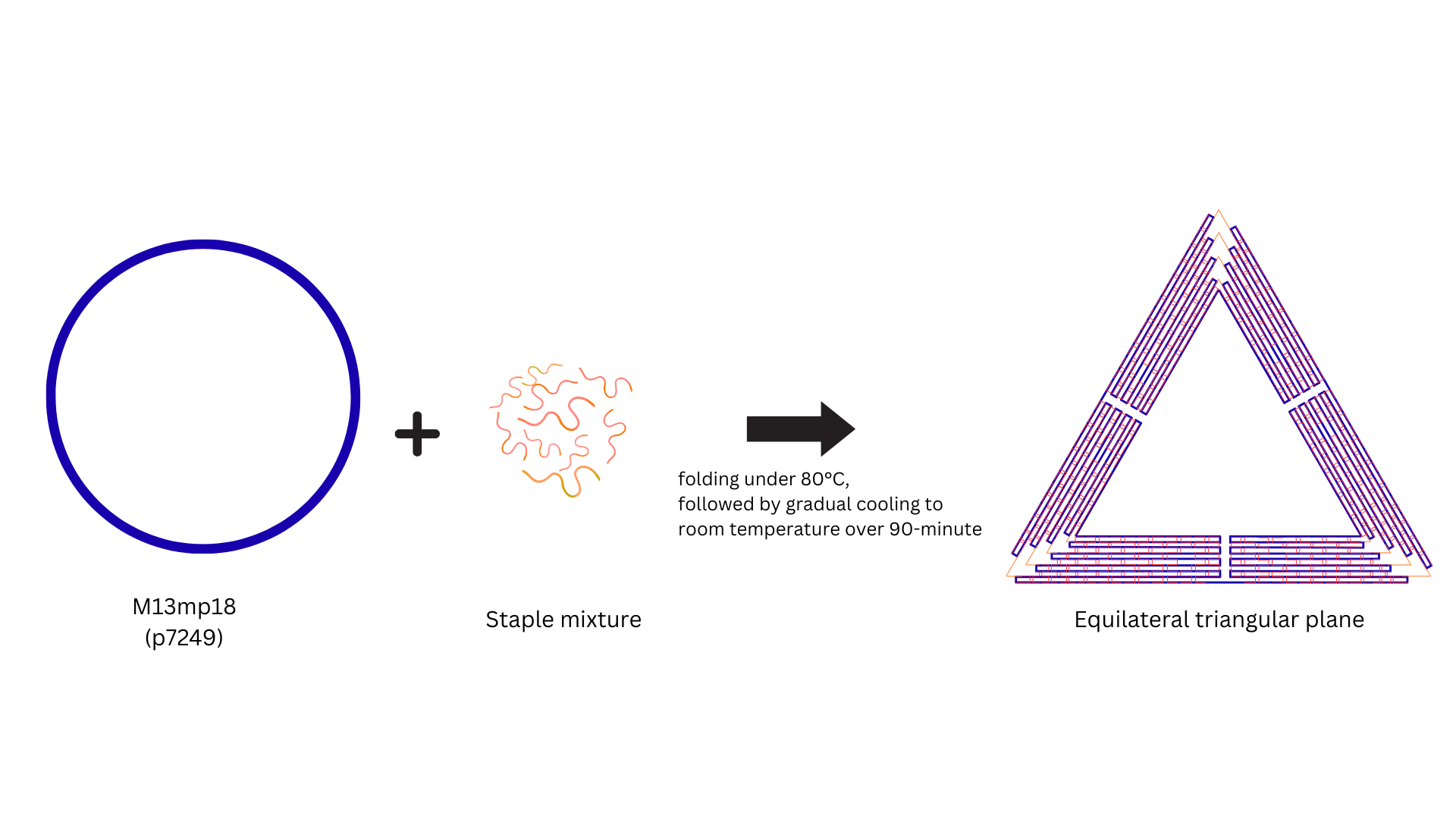
The M13mp18 (ssDNA, p7249) was used as the scaffold, and 200 staple strands were co-folded into a planer equilateral triangle (~ 120 nm). Experimental conditions were folding at 80°C, followed by gradual cooling to room temperature over 90 minutes.
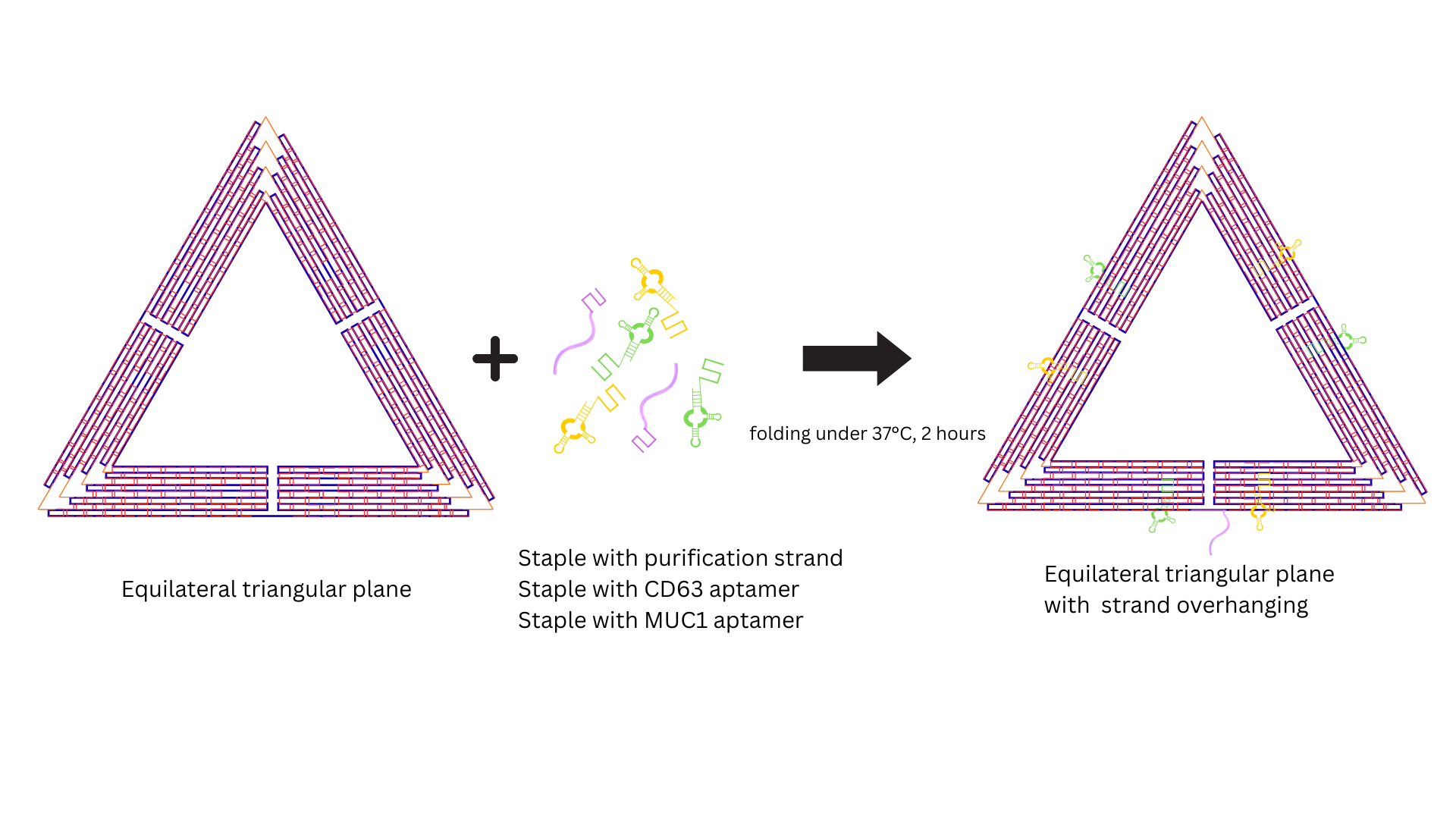
The folded DNA origami has a planar equilateral triangle structure. Then, place it under 37°C for 2 hours and attach the following aptamers: 1. Purification strand 2. CD63 aptamer 3. MUC1 aptamer.

Future

This research has completed the folding of a DNA origami and successfully linked it to a specific aptamer. Furthermore, magnetic bead purification was used to initially isolate the origami, facilitating subsequent exosome binding experiments. However, due to time constraints, subsequent experiments are still ongoing and have not been completed. Please see the following three points.
- Purify exosomes from NK92 cell culture medium.
- Establish a stable binding system between DNA origami and exosomes.
- Evaluate the specific binding ability and cytotoxicity of the combined complex to A549 lung cancer cells.
If the subsequent research can be successfully completed, it is expected that this innovative strategy can be further applied to other disease models to develop a nano-delivery platform with multi-targeting potential. DNA origami by modifying the structure externally, exosomes can be precisely targeted to different tissues and cells, exerting their biological delivery and immune regulation functions. Ultimately, this study hopes to establish a nanoscale delivery system with both targeted and immune regulation potential as the basis for future immune-related applications.